Abstract
Human cells acquire somatic mutations throughout life, some of which can drive clonal expansion. Such expansions are frequent in the hematopoietic system of healthy individuals and have been termed clonal hematopoiesis (CH). While CH predisposes to myeloid neoplasia and some non-hematological diseases, we have limited understanding of how and when CH develops, what factors govern its behavior, how it interacts with aging and how these variables relate to malignant progression.
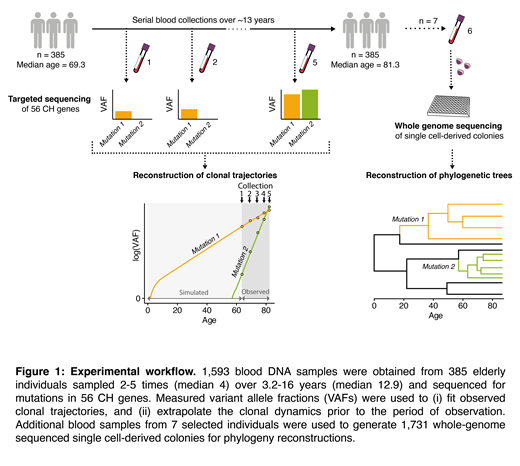
To address these questions, we adopted the experimental approach outlined in Figure 1. We began by analyzing 1,593 blood DNA samples from 385 elderly individuals, each sampled up to 5 times over ~13 years. We used deep targeted sequencing to identify and track 697 CH clones over this period. We constructed a logistic regression model and found that the great majority of clones (92.4%) expanded at a stable exponential rate during old age, enabling accurate prediction of future clonal growth (mean absolute error of measured vs predicted clone size: 3.5%). Different mutations drove substantially different growth rates, with mutations in DNMT3A and TP53 displaying the slowest average annual growth rates of ~5%. Clones with mutations in the other common driver genes (TET2, ASXL1, PPM1D and SF3B1), expanded at roughly twice this rate, i.e. ~10%/yr. The most rapidly expanding clones were those carrying SRSF2-P95H mutations, which grew at rates of over 50%/yr. Most hotspot mutations, including those at DNMT3A-R882, were not associated with faster growth than other mutations in the same gene. Growth of different clones bearing the same driver mutation varied by +/-5%/yr, proportionately impacting "slow" drivers most substantially.
To investigate lifelong clonal behavior, we first used retrograde extrapolation of observed trajectories to infer the age at which each clone had begun to expand. This led to projected ages at clonal foundation that preceded conception for many clones, implying that their early growth must have been faster than the rate we observed during old age. This was most striking for DNMT3A, for which more than two thirds of projections were implausible (i.e. onset pre-conception), less common for TET2 and very uncommon for splicing factor genes. Quantification of the minimum lifetime rate at which clones must have grown in order to reach observed VAFs confirmed that deceleration was particularly marked for DNMT3A-mutant clones, whose early growth was at least twice as fast as growth during old age.
To further characterize clonal dynamics over the human lifespan, we analysed hematopoietic phylogenies from 7 older individuals (aged 73-83 years), constructed using somatic mutations as lineage-tracing barcodes in 1,731 whole genome-sequenced hematopoietic colonies. We used the timing and density of clonal branching to age the onset of clonal expansions and to reconstruct their lifelong growth trajectories. In conjunction with findings from our longitudinal cohort, we revealed distinct patterns of clonal behavior. DNMT3A-mutant clones preferentially initiated and expanded early in life and displayed slower growth in old age, in the context of an increasingly competitive oligoclonal landscape. In contrast, TET2 mutations were able to found clones fairly uniformly through life and drove more stable growth across ages. This disparity in behavior might explain why TET2 "overtook" DNMT3A as the most common CH driver after the age of 75 years. In diametric contrast to DNMT3A and unlike other genes, CH driven by mutant U2AF1 and SRSF2-P95H only initiated late in life and exhibited the fastest expansion dynamics.
Finally, we explored the relationship between CH growth rates and malignant progression, building on our previous study of Acute Myeloid Leukemia (AML) risk prediction (Abelson et al, Nature 2018), and applying dN/dS analyses to published data (Papaemmanuil et al, Blood 2013 and NEJM 2016) to quantify selective pressures on different gene mutations in myeloid cancers. We found that mutations associated with faster CH growth were also those associated with higher risk of progression to AML (adjustedR 2=0.55, p=0.004) and were under the strongest selective pressure in AML and Myelodysplastic Syndromes (adjustedR 2=0.19, p=0.002).
Collectively, our findings characterize the lifelong natural history of CH and give fundamental insights into the interactions between somatic mutation, aging and clonal selection.
Campbell: Mu Genomics: Current holder of individual stocks in a privately-held company, Membership on an entity's Board of Directors or advisory committees. Vassiliou: Astrazeneca: Consultancy; STRM.BIO: Consultancy.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal